Practical Session 1: Bacteria Collection <<<
Practical Session 2: DNA Extraction
This training session has the aim that you create Experimental Steps to extract DNA from one existing Bacteria and create an Entry to summarize the Experiment.
In addition the aim is to link the Media created in Session 1 to one of the Experimental Steps and to share your Experiment with another user of the openbis instance. The shared user should comment on your Experiment via the Comment Log.
Please try to follow this list of tasks.
Create in a Test Project within your own user space:
Plasmid Collection (PLASMID)
DNA Collection (DNA)
Add 1 Plasmid to your Plasmid Collection
- Add 1 Bacteria which is the offspring (Child) of an existing Bacteria and carries the created Plasmid
Create in a Test Project within your own user space:
Default Experiment (Name: DNA Extraction)
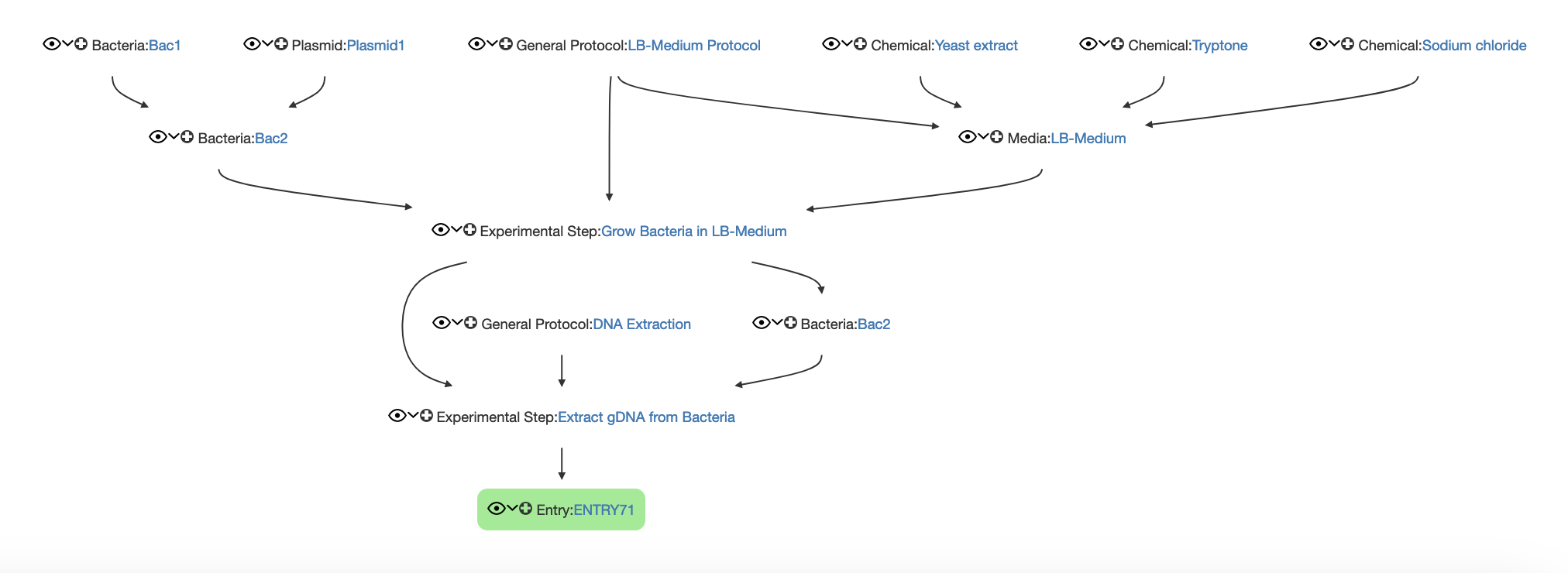
Create an Experimental Step 1 in the Default Experiment, which should describe growing a bacterial culture
- Link the LB-Media and LB-Media Protocol to this Experimental Step 1
- Create a Suspension Culture Protocol and link the protocol as Parent to the created Experimental Step 1
- Create a copy of the used Bacteria
- Link the copied Bacteria to Experimental Step 1 as Child
- Create a DNA extraction protocol in the Protocol Collection
- Create an Experimental Step 2 starting from Experimental Step 1, which should describe the DNA extraction
- Link the copy of the used Bacteria and the DNA extraction protocol as Parent
- Create a DNA entry and link it as Child to the Experimental Step 2
- Create an Entry starting from Experimental Step 2, which should describe the results of the experiment
- Add a gel electrophoresis photo (example gel photo) to the Rich Text Format Field
- View the Hierachy Graph of the Experimental Step 2
- Use the Manage access button of the Test Project to share it with another openbis user with the role User
- Ask the user that you give access rights to your Test Project to leave a comment via the Comment Log
Note: The other user needs to edit the corresponding Experimental Step to leace a comment and Save.
Note: This protocol is just an example and further downstream steps, like doing gel electrophoresis, PCR or sequencing can be added as additional Experimental Steps.
Resources
Plasmid
- Backbone: pGEX4T1
- Bacterial Antibiotic Resistance: bla
gDNA extraction kit
- H225-H315-H317-H318-H334-H335-H336-H410 (highly flammable liquid and vapour, causes skin irritation, may cause an allergic skin reaction, causes serious eye damage, may cause allergy or asthma symptoms or breathing difficulties if inhaled, may cause respiratory irritation, may cause drowsiness or dizziness, very toxic to aquatic life with long lasting effects)
- P280 (wear protective gloves/protective clothing/eye protection/face protection)
- Instruction: ROTI®Prep gDNA Mini 2.0
- Specification: ROTI®Prep gDNA Mini 2.0
Suspension Culture
- Prepare LB medium by weighing appropriate powder medium and adding to water in a sterile flask
- Autoclave the broth and cool to room temperature (Alternatively ready-to-use LB medium may be used)
- In a laminar flow chamber, transfer approximately 1 mL of overnight E. coli culture to the flask
- Seal the mouth of the flask with sterile cotton (non-absorbant) plugs; ensure the flask is not tightly sealed
- Incubate overnight at 37° C with continuous shaking
DNA Extraction Protocol
see section 4.2.1 DNA isolation from bacterial cell cultures Instruction: ROTI®Prep gDNA Mini 2.0
Next >>> Practical Session 3: PCR Protocol
© Kristian K Ullrich (2024) - ullrich@evolbio.mpg.de